page 2
(The Study of Threes)
http://threesology.org
Visitors as of 11/18/2019
| Pages in the Biological & Physiological 3s Series: | ||
| B/P 3s pg. 1 | B/P 3s pg. 2 | B/P 3s pg. 3 |
| B/P 3s pg. 4 | B/P 3s pg. 5 | B/P 3s pg. 6 |
| B/P 3s pg. 7 | B/P 3s pg. 8 | B/P 3s pg. 9 |
| B/P 3s pg. 10 | B/P 3s pg. 11 | The Devil's Advocate and 3s Research |
One of the central questions of current molecular biology is: "Which of the three fundamental biopolymers arose first?"
3 origin theories can be realized by consideration of the three biopolymers:
The dual and triple origin hypothesis suggest a simultaneous or (somewhat of a simultaneous) development that may have occurred in separate environments and somehow became mixed. Separate niche developments that fused into a single configuration (so to speak), would be an instance of a 3 to 1 ratio. An example of separate niche developments fused into a single configuration appears to be individual cells containing organelles such as the mitochondria, golgi complex, lysosome, etc. Each has a specialized function that work symbiotically (together). For additional related comments: http://www.panspermia.org/rnaworld.htm |

If we look at the number of strands of each of these, we find RNA is predominantly single stranded, DNA is predominantly double stranded, and Proteins may have a Primary ~ Secondary ~ Tertiary structure, with a composite called the Quaternary. Clearly, each of these has a "1" and a "2" in that we can find some RNA with a double strand and some DNA with a single strand. If the numerical quantity of strands is indicative of placement of arrival on the biological scene, then one might want to conclude that they may have all arrived, in some form, simultaneously.
Primordial soup biopolymer precursors |
RNA ↓ |
DNA ↓ |
Proteins ↓ |
|
| 6-5 billion years ago?* | Single strand | Yes | Yes | Yes |
| 5-4 billion years ago?* | Double strand | ?? | Yes | Yes |
| 4-3 billion years ago?* | Triple strand | ??? | ??? | Yes |
*These time periods are meant as "conversational" comparisons. No doubt some readers may find the time periods as being too "expansive" for the intellectual rigor they prefer. The fact remains that all estimates are guesstimations whether or not one attempts to use present day "evidence" to support their conclusions. For example, it is thought that the Earth is, at most, 4.567 billion years old. This being the case, the above numerical dates need only be substituted with the correct time periods... yet, this does not alter the overall 1 - 2 - 3 developmental formulation idea. Humorously, I think humanity arose from some alien's left-over sandwich they forgot while having a picnic on Earth many billions of years ago. Humanity began as a mold. This is the only way to explain why so many kids like peanut butter and jelly sandwiches. I think we should dub this the PB & J life origin theory. I should win the Nobel Prize for it. But no applause, please. Grant money will do just fine. Now where is that sandwich I absent-mindedly laid down while writing this? HMMM... Oh well, a new species might well be in the making... however, we might want to consider the possibility that the different races are due to different brands of peanut butter, different types of jam or jelly, and what type of bread was used. | ||||
However, if we consider that there is an environmental influence on biological development to follow a three- patterned course, then it is clear that RNA was too primitive to proceed to a full expression of a three-patterned structure. It is also clear that while DNA did achieve a full 2-patterned expression of development because it is predominantly double stranded, it too is primitive in the sense it did not achieve a full 3-patterned expression in the number of strands. The full expression of a "3" was achieved by the 3-stranded Protein.
The initial development of all three substances may have occurred in an overlapping fashion with RNA coming first, DNA coming second, and Proteins coming third in the sense of the ability to proceed further in a 1~ 2~ 3 development, and not in the sense of a chicken versus egg controversy. Nonetheless, we might want to consider that Proteins came first and it is because of its pristine arrival that it was able to acquire the full measure of a 1-2-3 developmental plan that was available long ago, but was not available for DNA which was influenced by a 2-patterned environmental influence and RNA which arrived when environmental circumstances only offered a single format floor plan. In other words, there once existed a three-patterned environmental circumstance which influenced a three-patterned floor plan for proteins, but that on the arrival of DNA, only a two-patterned environmental influence which was then followed by a one-patterned environmental influence, which would offer an explanation of why we can find double and single stranded forms of RNA and DNA. Yet the fact that DNA makes RNA makes Protein, may suggest that some form of DNA did come first:
http://www.nobel.se/medicine/educational/poster/1993/dna-rna.html
However, the occurrence of an overlapping development suggests a necessity of arrangement during a specific period under particular circumstances that we might be able to recreate in a controlled setting. A simultaneity of development with an observable progressive complexity might be unraveled to expose an underlying simplicity that must occur in a simultaneous and overlapping fashion in order for the progressive design of complexity to develop (as a sort of "emergent" property). In some fashion, this could be equated with a punctuated equilibrium, even though such an "equilibrium" must have its own developmental progressiveness.
Reflectively speaking, it must also be considered that biological development circumstances will change (is changing), with the present DNA-RNA-Protein complexity little more than a pristine expression of something yet more complex to evolve.
| RNA | DNA | Proteins | |
| Single stranded = | MAJOR | minor | Simple |
| Double stranded = | minor | MAJOR | Intermediate |
| Triple stranded = | no | See article below concerning single stranded RNA and triple stranded DNA | MAJOR (complex) |
| 3 to/in 1 array = | A-C-G + Uracil | A-C-G + Thymine | Si-In-Co |
| (By combining the Simple + Intermediate + Complex protein structures, this 3-into-1 formula is called a quaternary.) |
3 polymers each made
up of repeating units of subunits called monomers.
- 1 of 3. Proteins, function = enzymes (biological catalysts)/metabolism, transport of molecules, DNA & RNA synthesis, structure of cell.
- 2 of 3. Polysaccharides, function = energy source.
- 3 of 3. Nucleic Acids,
function = storage and expression of genetic material.
- 1 non-polymer (not made up of smaller
subunits).
1 of 1. Lipids, Phospholipids = made up of fatty acids, glycerol, phosphate and are the main structural component of cell membranes. (Cell membrane is a lipid bilayer.)
http://www.calpoly.edu/~selrod/Lecture02biochemanat.html
AFM of single-stranded RNA, triple-stranded DNA and helix turns http://www.physics.ucsb.edu/~hhansma/helix-ss.htm Single-stranded RNA 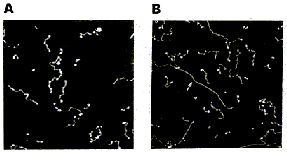 Single-stranded polyA (A, B) differs from double-stranded DNA mainly in its persistence length. The persistence length of ss polyA is ca. 40 nm, while the persistence length of dsDNA under the same conditions is ca. 80 nm. PolyA in water also protonates to form a very stiff double helix with a persistence length of ca. 600 nm, (e.g., arrow in B) Images are 1 mm2. (From (1).) Triple-stranded DNA 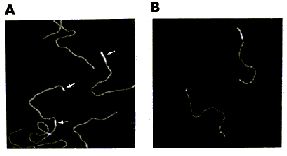 Features that appear to be triple-stranded DNA can be seen on poly(dA):poly(dT) (A, B) and poly(dG):poly(dC). The putative triple-stranded DNA is ca. twice as high as dsDNA. Images are 1 mm2. (From (1).) Helix turns on dsDNA (2) 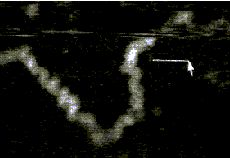 The best images of helix turns with tapping AFM show more detail than images of helix turns with contact AFM (3, 4). These helix turns were imaged in propanol with an unusually good tip and cannot be obtained reliably. Scale bar is 10 nm. Shao's group can see helix turns on dsDNA much more reproducibly but in less detail than the above image, using samples of densely packed DNA on a positively charged lipid bilayer, imaged with contact AFM (3). We have not yet been able to image such a sample with tapping AFM to see if the helix turns can be resolved in more detail. The DNA in propanol shows intriguing deformations of the double helix where the DNA bends sharply that would be fascinating to image more. We believe that only the major groove was detected, because the periodicities are 3-4 nm. Densely packed DNA on lipid should be better for getting high resolution, because there is less interaction between the tip and the DNA than with isolated DNA molecules (5). References: 1. Hansma, H. G., I. Revenko, K. Kim, and D. E. Laney. 1996. Atomic force microscopy of long and short double-stranded, single-stranded and triple-stranded nucleic acids. Nucleic Acids Res. 24:713-720. 2. Hansma, H. G., M. Bezanilla, D. L. Laney, R. L. Sinsheimer, and P. K. Hansma. 1995. Applications for Atomic Force Microscopy of DNA. Biophys. J. 68:1672-1677. 3. Mou, J., D. M. Czajkowsky, Y. Zhang, and Z. Shao. 1995. High-resolution atomic-force microscopy of DNA: the pitch of the double helix. FEBS Lett. 371:279-282. 4. Hansma, H. G., and P. K. Hansma. 1993. Potential applications of atomic force microscopy of DNA to the human genome project. Proc. SPIE - Int. Soc. Opt. Eng. (USA). 1891:66-70. 5. Hansma, H. G. 1995. Polysaccharide helices in the atomic force microscope. Biophys. J. 68:3-4. |
3-stranded DNA helix was initially offered by James Watson and Francis Crick in 1951. It was copied from ideas based on molecular structures developed by Linus Pauling. It was Rosalind Franklin who initially offered the correct design of a 2-stranded form that she was never given credit for until the 21st century. The efforts and contributions of women are frequently overlooked.
3-part Linus Pauling combination method (that he used to study proteins) that was adopted by Watson and Crick in their research on DNA:
- Model-building.
- Chemical knowledge
- Modern physics.
3 referenced ideas which assisted James Watson and Francis Crick in winning the Nobel Prize for unraveling the DNA structure:
- Idea of base pairing.
- Idea of double-stranded helix.
- Two photos numbered 51 and 52.
See articles at:
http://osulibrary.orst.edu/specialcollections/coll/pauling/dna/narrative/page1.html
3-dimensional complex protein structures can be described by 3 + 3 (six) levels of structural organization: Primary, Secondary, Super- secondary, Domain, Tertiary and Quaternary.
3 class grouping for nearly all naturally occurring protein fibers according to their X-ray diffraction patterns:
Alpha-type containing (among others) the proteins of unstretched hair, fingernail, horn and bacterial flagella.
Beta-type characterized by stretched hair and silk fibroin.
Gamma-type containing the protein collagen.
http://broccoli.mfn.ki.se/pps_course_96/ss_960723_2.html
| 3 main components (Triad) of the Biosphere: RNA~ DNA~ Proteins | |
RNA:
Predominantly single stranded (Image source: Revelations of a biological abacus) | |
DNA: Predominantly double-stranded (Double
Helix) |
 DNA Triple Helix http://www.cryst.bbk.ac.uk/BBS/whatis/DNAtriple_gr.html |
Proteins: Predominant triple conformation
structure | |
| 3 to 1 ratio: {Adenine~ Cytosine~ Guanine} + Uracil = RNA
3 to 1 ratio: {Adenine~ Cytosine~ Guanine} + Thymine = DNA (Adenine + Thymine) = 2 hydrogen bonds (Guanine + Cytosine) = 3 hydrogen bonds (???) + (???) = 1 hydrogen bond??? | |
3 natural forms of DNA: (A ~ B ~ Z), whose origins are related to the conformation of the sugar 2'-endo/ C 3'-endo and the orientation of the base relative to the sugar (syn/anti):
The B-form is the common natural form, prevailing under physiological conditions of low ionic strength and high degree of hydration. B-DNA arranges 10 nucleotides per helix tour, all of conformation C2'-endo/anti. |
The A-form is sometimes found in some parts of natural DNA in the presence of a high concentration of cations or at a lower degree of hydration (<65%). A-DNA possesses 11 nucleotides per tour (all C3'-endo/anti and two grooves (a narrow deep major and a wide shallow minor). The Z-form (Zigzag chain) is observed in the DNA G-C rich local region. Z-DNA is longer, thinner and has an unusual left-handed helix of 12 bases pairs/tour) with a single narrow deep groove. The Zigzag form mainly results from the alternation of purines C3'-endo/syn and pyrimidines C2'-endo/anti. |
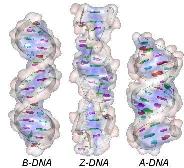 --- DNA --- The C- and D-forms are unusual subclasses of the B-type. C-DNA is sometimes observed under 45% of hydration while D-DNA is only found in artificial DNA. |
http://www.geneticengineering.org/chemis/Chemis-NucleicAcid/DNA.htm
Note: For those who are numerologically inclined, the values of 10, 11, and 12 in the previous table can be seen as a 1~ 2~ 3 representation:
11: 1 + 1 = 2
12: 1 + 2 = 3
3 distinct (classically described) steps to RNA Transcription: Initiation, Elongation and Termination.
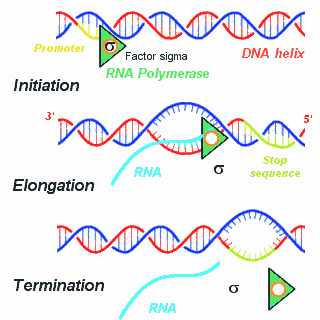
Initiation occurs when the RNA polymerase holoenzyme binds at a special sequence in DNA called a "promoter." The promoter consists of consensus sequences containing specific strings like TATA (Pribnow box) and CAAT (in eukaryotes). An additional small protein, the "factor sigma," attaches to the polymerase and stabilizes it, locking it on the DNA strand to be transcripted. Then, the polymerase separates the double stranded DNA to form a bubble allowing the first nucleoside triphosphate to pair with the complementary DNA nucleotide.
Elongation of the RNA chain involves successive additions of nucleotides in the 5' to 3' direction.
Termination occurs when a Stop signals indicating the end of the gene is encountered. The termination signal is generally a GC-rich palindrome forming a local stem-loop structure in the RNA , followed by an oligo A region. This sequence disrupts the base pairing of newly synthesized RNA with the DNA template, forcing the RNA and the polymerase to fall off. Sometimes termination also involves a specific protein (Rho protein).
http://www.geneticengineering.org/chemis/Chemis-NucleicAcid/RNA.htm
| Amino Acid | 3-lettered abbreviation | 1-lettered abbreviation |
| Alanine | Ala | A |
| Arginine | Arg | R |
| Asparagine | Asn | N |
| Aspartic acid | Asp | D |
| Cysteine | Cys | C |
| Glutamic acid | Glu | E |
| Glutamine | Gln | Q |
| Glycine | Gly | G |
| Histidine | His | H |
| Isoleucine | Ile | I |
| Leucine | Leu | L |
| Lysine | Lys | K |
| Methionine | Met | M |
| Phenylalanine | Phe | F |
| Proline | Pro | P |
| Serine | Ser | S |
| Threonine | Thr | T |
| Tyrosine | Tyr | Y |
| Tryptophan | Trp | W |
(In the above table, there are seven letters not being used: B,J,O,U,V,X,Z).
The symbol chosen for an amino acid is derived from its trivial name, and is usually the first three letters of this name. It is written as one capital letter followed by two lower-case letters, e.g. Gln (not GLN or gln), regardless of its position in a sentence or structure. If any other convention is used in representing residues, e.g. to emphasize homology, this should be stated clearly whenever it is used. When the symbol is used for a purpose other than representing an amino-acid residue, e.g. to designate a genetic factor, three lower-case italic letters may be used, e.g. gln.
http://www.chem.qmul.ac.uk/iupac/AminoAcid/A1416.html
There are difficulties in using the three-letter system in presenting long protein sequences. A one-letter code is much more concise, and is helpful in summarizing large amounts of data, in aligning and comparing homologous sequences, and in computer techniques for these processes. It may also be used to label residues in three-dimensional pictures of protein molecules.
The one-letter system is less easily understood than the three-letter system by those not familiar with it, so it should not be used in simple text or in reporting experimental details of sequence determination. It is therefore recommended for comparisons of long sequences in tables and lists, and in other special uses where brevity is important. If both it and the single-letter system for nucleotide sequences are used in the same paper, particular care should be taken to avoid confusion.
http://www.chem.qmul.ac.uk/iupac/AminoAcid/A2021.html
3 (gases) to 1 (water) ratio for Stanley Miller's
Chemical Evolution experiment:
(Perhaps a type of three-electrode system should have been tried?... such a "3-electrode" system was used with the advent of the Earth's rotation slowing which created a Dawn - Noon- Dusk [Triple- patterned] stroboscopic irradiation. See the following page for comments on the Earth's rotation:
http://www.threesology.org/rotation-rate-1.php)
In the following example, I have placed Stanley Miller's Experiment next to a 3-electrode system configuration so you can make a visual comparison.
Stanley Miller's Experiment
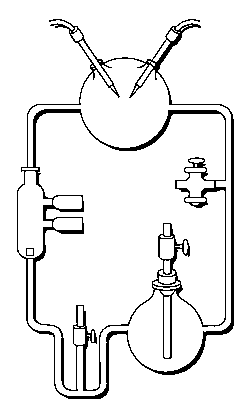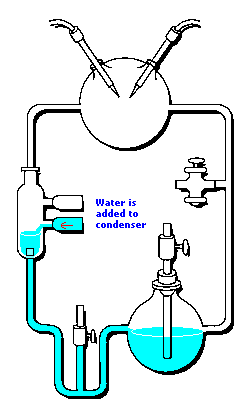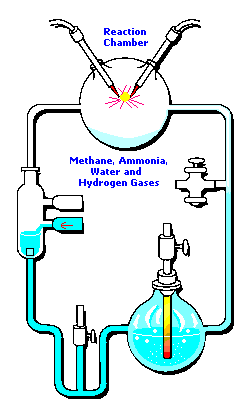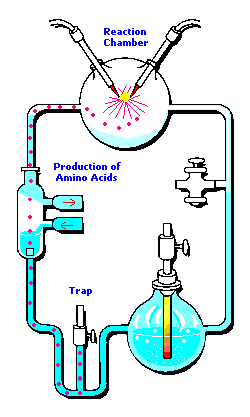
|
Three-electrode example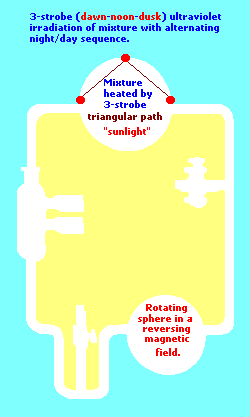
|
| ---EXOBIOLOGY:
Stanley Miller--- http://www.accessexcellence.org/WN/NM/miller.html | |
Experiments in chemical evolution are focused on a variety of interpretations related to an early Earth environment. One of my own interpretations has been from the perspective of speculating on the possibility that the triplet (codon) pattern and the helical structure of DNA might be representations of patterns which were present so many billions of years ago, if in fact they do not now exist in some fashion. (In other words, the DNA strand may exhibit an environmentally influenced biological rhythm. However, the initial environmental influence may not now exist at all or in the form which originally influenced the "coiled" formation in DNA.)
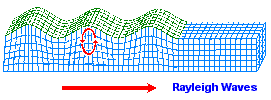
While most images of the double helix DNA are presented to viewers in a vertical (up/down) position, on occasion you might come across one that is placed horizontally. The horizontally placed image reminds me of ocean waves. While some chemical evolution experiments have relied on static environments, it is of value for experimenters to adopt more dynamical environmental scenarios other than the simple application of "movement" in the sense of irradiation from sunlight, electrical impulses, or volcanic-like heat sources. If we strip away the labeling associated with one of the above portrayals of the DNA strand and then compare it to waves (whether of typical day-to-day oceanic patterns or patterns found in earthquakes such as the images below indicate, there are some striking similarities. Such similarities also influences us to consider whether or not they are suggestive of the depth(s) that the early life processes may have begun at. (However, I am also reminded of sound wave patterns:)
Perhaps one could offer the amusing scenario that life on Earth began by being seeded by an alien(s) who irradiated a portion of the ocean with a particular sound wave(s). Maybe some alien rock, rap, or jazz group used to practice on Earth because their parents didn't like the kind and level of noise enjoyed by a younger generation. HA!

 |
 |
 |
 |
Earthquake images adapted from:
http://www.allshookup.org/quakes/wavetype.htm

3 component nucleotide: Nitrogenous base~ Pentose sugar ~ Phosphate group
{Nucleotides typically have 1~ 2~ (or) 3 phosphate groups: Monophosphate ~ Diphosphate ~ Triphosphate}
3 ionized forms of amino acids: Nonpolar ~ Polar ~ Electrically charged
- 3 Billion base pairs: common reference to the human genome
- 3 basic units to life: Atoms ~ Molecules ~ Cells
- 3 ERR subtypes in protein-based assays: Alpha ~ Beta ~ Gamma
- 3 cell membrane theories: (1st) Monolayer ~ (2nd) Bi-lipid layer ~ (3rd) Trilaminar
- 3 common references to human body cell quantity: 1 trillion ~ More than a trillion ~ 100 trillion
- 3-stranded Collagen is considered most abundant protein in our body (Triple Helix)
Three major ways collagen fibers are organized:
- By orientation of the cell that secreted them.
- By association with the underlying ECM (extra cellular membrane).
- By activity of fibril-associated collagens.
3 to 1 ratio: 99% of all living systems are made up
of Hydrogen ~ Oxygen ~ Nitrogen + Carbon
(if we include sulfur and phosphorus, we
have a 3 + 3 arrangement).
Tribonucleic acid (TNA), consisting of a hemiactal backbone of surface-anchored phosophotrioses, has been proposed as a hypothetical precursor to RNA and DNA, by G. Wachtershauser, 1988.
Three modes of association between metal complexes and DNA are generally distinguished:
External binding 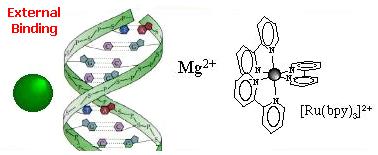 External association of the complex in the atmosphere of ions of the DNA polyelectrolyte... ...This
association is electrostatic in nature (Ru(II) complexes are 2+ positively
charged and the DNA phosphate sugar backbone is negatively charged). This
association mode was proposed for [Ru(BPY)3]2+ as the luminescence enhancement
of this complex upon binding to DNA is strongly dependent on the ionic
strength. Cations as Mg2+ usually interacts also in this way. Groove binding 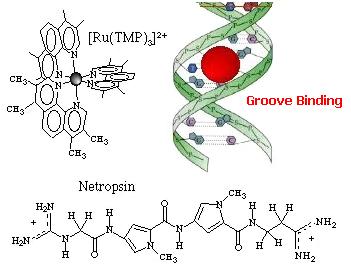 Adsorption of the complex in the DNA grooves. The molecules approaches within van der
Waals contact and resides in the DNA groove. Hydrophobic and/or
hydrogen-bonding are usually important components of this binding process, and
provide stabilization. The antibiotic netropsin is a model groove-binder
(to see the X-ray structure of this groove-binder click on the molecule).
Geometric and steric factors also play a role as shown with [Ru(TMP)3]2+ (TMP =
3,4,7,8-tetramethyl phenanthroline) where the methyl groups prevent
intercalation. Intercalation 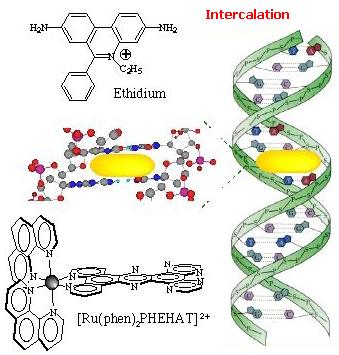 Intercalation of a planar ligand of the complex in the DNA base pairs stack. This association involves the insertion of a planar fused aromatic ring system between the DNA base pairs, leading to significant p-electron overlap. This mode of binding is stabilized by stacking interactions and is thus less sensitive to ionic strength relative to the two other binding modes. This mode of binding is usually favoured by the presence of an extended fused aromatic ligand as PHEHAT [1] or DPPZ [2]. Indeed with less extended aromatic systems, the intercalation is usually prevented through clashing of the ancillary ligands with the phosphodiester backbone, so that only partial intercalation can occur as it is the case for [Ru(phen)3]2+ [3]. http://www.photobiology.com/photoiupac2000/pierard/Interactionmain.html |
3 exon distinction: Findings are presented concerning genetic information conservation between the glucocorticoid response element (GRE) DNA and the c-DNA encoding the glucocorticoid receptor (GR) DNA binding domain (DBD). The regions of nucleotide subsequence similarity to the GRE in the GR DBD occur specifically at nucleotide sequences on the ends of exons 3, 4 and 5 at their splice junction sites. These sequences encode the DNA recognition helix on exon 3, a beta strand on exon 4, and a putative alpha helix on exon 5, respectively. The nucleotide sequence of exon 5 that encodes the putative alpha helix located on the carboxyl terminus of the GR DBD shares sequence similarity with the flanking nucleotide regions of the GRE.
http://www.msi.umn.edu/Projects/mg90601/pnas1.html
Updated Posting: 06-Aug-2017... 08:18 AM
Latest Update: Sunday, 15th September 2019... 1:36 PM
Latest Update: Friday, February 24th, 2023... 11:17 AM
Herb O. Buckland
herbobuckland@hotmail.com