page 4
(Using a philosophy of "threes" as a tool
for dissection and comparative analysis)
http://threesology.org
Anat., Bio. Phys. Researchers as of 3/17/2021
Pages in this series:
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
Using a "threes analysis tool" when studying anatomy, biology and physiology is particularly easy since there are multiple representative examples, and made more so if we adopt a larger holistic philosophy which takes into account the existence of a 1, 2, 3 sequencing development that in some cases we may define as a maturational scenario involving occasions of fusion. No less, we must apply the philosophy across the spectrum of representative examples and not let our conventional labels create barriers to comparison. We must also view our growing list of examples from multiple venues of information as a inter-linked three (or more) multi-dimensional grid instead of looking at everything in a two-dimensional format. Relevant connections may escape our considerations if we don't apply an expansive perception.
For example, if we look solely (as an isolated model) at the single stranded RNA in terms of an "RNA World" hypothesis and not view it as a numerically expressive segment of a larger developmental scenario followed by a double-stranded DNA, the dissection of both may or may not lead us to identify the presence of a 3 to 1 ratio:
| 3 main components (Triad) of the Biosphere: RNA~ DNA~ Proteins |
|
RNA: Predominantly single stranded3 main types: Messenger (mRNA)- Ribosomal (rRNA)- Transfer tRNA), (and "small nuclear" is sometimes included as an additional [3 to 1] type) 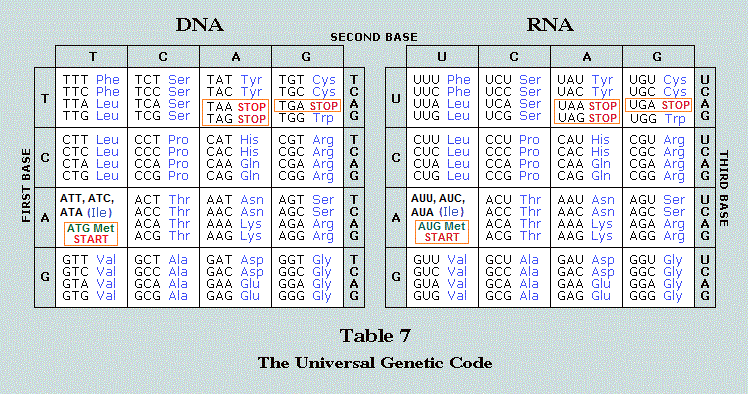 Types of small RNA Revelations of a biological abacus) 3-site tRNA model is well established, 2-site model is dead |
DNA: Predominantly double-stranded (Double
Helix)  DNA Triple Helix |
Three noted forms of DNA: As far as we know, or at least have labeled our current understanding of genetics, DNA and RNA are the two nucleic acids which are recognized during this period in humanity's level and type of biological investigation. Also, Depending on how one wishes to classify DNA a two-patterned model may be preferred. "There are two major types of DNA- Genomic DNA and Mitochondrial DNA" 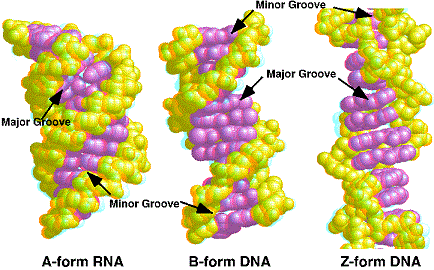 Biochemistry Topics Types of DNA and RNA It should be of interest for those who like to comparative wide swaths of information from different subject areas, that the lower values of numbers are used again and again, like an infant that is babbling with "reduplications". One must wonder if the mind of humanity is in a stuttering mode of operation need for survival in an incrementally deteriorating Sun-Earth-Moon complex? |
Proteins: Predominant triple conformation
structure |
 3 to 1 ratio: {Adenine~ Cytosine~ Guanine} + Uracil = RNA
|
3 natural forms of DNA: (A ~ B ~ Z), whose origins are related to the conformation of the sugar 2'-endo/ C 3'-endo and the orientation of the base relative to the sugar (syn/anti):
If the early Earth environment had a high incidence of Ultra-violet radiation, we must consider whether this was good or bad for early life processes. If it were bad, this would then suggest that RNA (or whatever occurred first), would have either been partially or totally immune to the effects. In other words, it would have had a tough "hide" that could not be easily branded by the hot sun. Was the single-stranded RNA more susceptible to mutation while the double-stranded DNA, because of its redundancy, created a more difficult situation for mutagenic appearances? Or does a double strand allow for greater "mixing" variability due to redundancy? However, let us not forget that mutations can and do appear, which permits adaptation to environmental changes. Will increased effects of the Sun due to "global warming" cause an increase in mutations... whether or not we are conscious of them because we are only looking for obvious indications and not subtle ones?
First, let's look at some basic information about mutations:
| The causes of mutations Mutations happen for several reasons:
Most of the mutations that we think matter to evolution are "naturally-occurring." For example, when a cell divides, it makes a copy of its DNA and sometimes the copy is not quite perfect. That small difference from the original DNA sequence is a mutation. 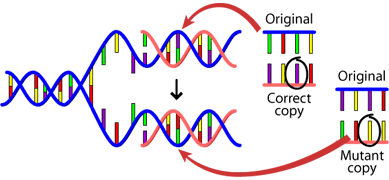
Mutations can also be caused by exposure to specific chemicals or radiation. These agents cause the DNA to break down. This is not necessarily unnatural — even in the most isolated and pristine environments, DNA breaks down. Nevertheless, when the cell repairs the DNA, it might not do a perfect job of the repair. So the cell would end up with DNA slightly different than the original DNA and hence, a mutation The only mutations that matter to large-scale evolution are those that can be passed on to offspring. These occur in reproductive cells like eggs and sperm and are called germ line mutations. A single germ line mutation can have a range of effects:
Some mutations don't have any noticeable effect on the phenotype of an organism. This can happen in many situations: perhaps the mutation occurs in a stretch of DNA with no function, or perhaps the mutation occurs in a protein-coding region, but ends up not affecting the amino acid sequence of the protein. 
A single mutation caused this cat's ears to curl backwards slightly.
Some really important phenotypic changes, like DDT resistance in insects are sometimes caused by single mutations. A single mutation can also have strong negative effects for the organism. Mutations that cause the death of an organism are called lethals and it doesn't get more negative than that. There are some sorts of changes that a single mutation, or even a lot of mutations, could not cause. Neither mutations nor wishful thinking will make pigs have wings; only pop culture could have created Teenage Mutant Ninja Turtles mutations could not have done it. |
Here's a brief reference to the RNA world hypothesis:
The RNA World and the Origins of Life To fully understand the processes occurring in present-day living cells, we need to consider how they arose in evolution. The most fundamental of all such problems is the expression of hereditary information, which today requires extraordinarily complex machinery and proceeds from DNA to protein through an RNA intermediate. How did this machinery arise? One view is that an RNA world existed on Earth before modern cells arose. 
Image From: The RNA World and the Origins of Life  Cover of Molecular Biology of the Cell
Cover of Molecular Biology of the CellMolecular Biology of the Cell. 4th edition. Alberts B, Johnson A, Lewis J, et al. New York: Garland Science; 2002. Copyright © 2002, Bruce Alberts, Alexander Johnson, Julian Lewis, Martin Raff, Keith Roberts, and Peter Walter; Copyright © 1983, 1989, 1994, Bruce Alberts, Dennis Bray, Julian Lewis, Martin Raff, Keith Roberts, and James D. Watson. NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health. According to this hypothesis, RNA stored both genetic information and catalyzed the chemical reactions in primitive cells. Only later in evolutionary time did DNA take over as the genetic material and proteins become the major catalyst and structural component of cells. If this idea is correct, then the transition out of the RNA world was never complete; as we have seen in this chapter, RNA still catalyzes several fundamental reactions in modern-day cells, which can be viewed as molecular fossils of an earlier world. |
Second, let's look at an excerpt of some information about specific RNA responsiveness to Ultra-Violet radiation,
in order to get some idea of how RNA interacts:
(Though we don't actually know the exact nature of the RNA which existed billions of years ago nor the intensity of the Sun's effects on the early
Earth's environment. We simply make educated guesstimations and create experiments like Stanley Miller and Urey.)
Miller-Urey Bong, 2010
Ascensión Ariza-Mateos, Samuel Prieto-Vega, Rosa Díaz-Toledano, Alex Birk, Hazel Szeto, Ignacio Mena, Alfredo Berzal-Herranz, Jordi Gómez
Nucleic Acids Research, Volume 40, Issue 4, 1
February 2012, Pages 1748 - 1766
https://doi.org/10.1093/nar/gkr822
Published: 11 October 2011
Abstract
A novel UV-C-light-induced ribozyme activity was discovered within the highly structured 5'-genomic regions of both Hepatitis C Virus (HCV) and the related Classic Swine Fever Virus (CSFV). Cleavage is mediated by exposure to UV-C light but not by exogenous oxygen radicals. It is also very selective, occurring at base positions HCV C 79 and CSFV A 45 in some molecules and at the immediately adjacent 5'-positions HCV U 78 and CSFV U 44 in others. Among other reaction products, the majority of biochemically active products detected contained 3'-phosphate and 5'-phosphate-end groups at the newly generated termini, along with a much lower amount of 3'-hydroxyl end group. While preservation of an E-loop RNA structure in the vicinity of the cleavage site was a requisite for HCV RNA self-cleavage, this was not the case for CSFV RNA. The short size of the reactive domains (~33nt), which are compatible with primitive RNA motifs, and the lack of sequence homology, indicate that as-yet unidentified UV-activated ribozymes are likely to be found throughout structured RNAs, thereby providing clues to whether early RNA self-cleavage events were mediated by photosensitive RNA structures.
Introduction
A significant number of novel functions, many of which have proven to be important in the study of gene function and expression, have been attributed to RNA. This, in turn, has led to speculation among experts regarding the evolutionary role of RNA in early biological systems. Most of these non-canonical functions for RNA molecules take place in specific regions containing higher order and evolutionarily conserved structural elements. NMR and X-ray crystallographic analyses have provided invaluable insight into some of these tertiary structures and, in the case of certain ribozymes, have revealed their active sites. One class of RNA that is particularly rich in non-Watson:Crick interactions is the Internal Ribosome Entry Site (IRES) domain located at the 5'-ends of several single-stranded RNA viruses such as Hepatitis C Virus (HCV) and the related animal pestivirus Classical Swine Fever Virus (CSFV) ( Figure 1 ).
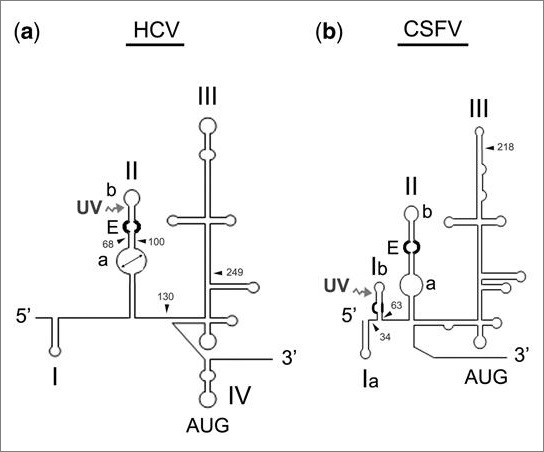
Secondary structure diagrams of the 5'-terminal regions of HCV and CSFV RNAs. Structural organization of the 5'-untranslated region (~400 bases long) of HCV and CSFV RNAs. The main differences between the two viral RNAs are that HCV lacks stem loop 1b of pestiviruses, and domain II is slightly longer in HCV than in the pestiviruses. A remarkable similarity is the presence of an E-loop structural motif in the stem loop IIb. ( a ) Bases 1 402 of HCV RNA; ( b ) Bases 1 380 of CSFV RNA. Each RNA's 5'-end is on the left; major common structural domains are depicted as domains I IV in HCV and I to III in CSFV. Domain I ('I') is bipartite in CSFV ('Ia' and 'Ib'). A UV-crosslinkable element previously described in HCV RNA is marked with a double arrowhead in stem loop IIa. A characterized E-loop in the immediate vicinity, as well as an analogous motif at the same site in CSFV RNA, is drawn in bold. An uncharacterized internal loop in stem loop Ib of CSFV RNA is also in bold. The position of AUG is indicated. Arrowheads locate the different transcripts size: 1'130, 1'249, 1 402 and 68 100 for HCV RNA and 1 218 and 34 63 for CSFV RNA. Cleavage site is indicated with an undulate arrow.
It is known that both DNA and RNA chains are affected by direct exposure to ultraviolet (UV) light, which in the case of DNA can result in the formation of pyrimidine dimers or random breaks. The effects of UV light on RNA are relatively unknown, although UV-C irradiation has been shown to induce mainly pyrimidine dimers and uridine hydrates and, less frequently, unspecific chain breaks. Base-radicals are major reactive intermediates of RNA UV-mediated degradation. Using an oligoribonucleotide with a position-induced base–radical model, the mechanism of RNA oxidative degradation has recently been elucidated in two consecutive studies. These studies indicated that the cleavage occurred internal to the initial radical generation site, referred to as intra-nucleotidyl strand scission, and at the adjacent 5' nt, referred to as inter-nucleotidyl strand scission, as a consequence of migration of the radical from its original base to the upstream base. Nevertheless, a part of unspecific degradation it should be emphasized that UV-C can also induce specific covalent bonds between bases in certain RNA regions containing structural motifs known as E-loops, an effect which has been utilized for the structural study of RNAs, including HCV RNA.
The biochemical mechanisms employed by RNA are surprisingly diverse. Nevertheless, the known chemistry promoted by natural RNAs alone is limited to transesterification and phosphate-bond hydrolysis, grouped together as nucleophilic cleavage reactions, or their reverse ligation reactions. One interesting exception, albeit in a DNA model, is the known ability of synthetic DNAs to promote self-repair of crosslinked thymine bases in a UV-dependent manner. It has been shown that a structural element involving four deoxy-G-residues absorbs UV-light activating crosslink repair.
Whilst performing UV-C irradiation experiments on HCV RNA transcripts, we found that, somewhat surprisingly, UV-C site-selectively cleaved HCV genomic RNA in the presence of Mg2+. We report here that UV-induced cleavage corresponds to a self-cleavage event specifically promoted by UV irradiation at a very specific location of HCV RNA. We also report the subset of key experiments which indicate that self-cleavage also occurs in the RNA of CSFV, thus making UV-C-induced self-cleavage a putatively general property of RNA.
This is the first evidence of a structure-dependent UV-activated selective self-cleavage of RNA in the absence of a photosensitizer.
Third, let's now move on to some structural elements of cells:
With respect to cells and the differences between animals and plants, we can begin with generalities and move on to more complex issues, without attempting to foray too deeply into cytological considerations which present specialized research interests. In other words, for the present course of presenting generalities of design, one key difference is to point out the presence of triplet microtubules in the animal cell because of the important role the structure plays.
(A microtubule is a) tubular structure of indefinite length, constructed from globular proteins called tubulins, which are found only in eukaryotic cells. Microtubules have several functions. For example, they provide the rigid, organized components of the Cytoskeleton that give shape to many cells, and they are major components of cilia and flagella (cellular locomotory projections). They participate in the formation of the spindle during cell division (mitosis). They also assist the movement of organelles (e.g., mitochondria), as well as the movement of vesicles from the cell bodies of neurons to the axonal tips and back to the cell bodies. Source: "Microtubule." Encyclopædia Britannica Ultimate Reference Suite, 2013.Centriole A morphologically complex cellular organelle at the focus of centrosomes in animal cells and some lower plant cells. Prokaryotes, some lower animal cells, higher plant cells, and a few exceptional higher animal cells do not have centrioles in their centrosomes. Centrioles typically are not found singly; the centrosome of higher animal cells contains a pair of centrioles (together called the diplosome), arranged at right angles to each other and separated by a distance ranging from 250 nanometers to several micrometers. Centrioles are typically 300- 700 nm in length and 250 nm in diameter. Although they can be detected by the light microscope, an electron microscope is required to resolve their substructure. At the electron microscopic level, a centriole consists of a hollow cylinder of nine triplet microtubules in a pinwheel arrangement (see illustration). Within each triplet, one microtubule (the A tubule) is a complete microtubule, while the others (the B and C tubules) share a portion of their wall with the adjacent tubule. In some cells these nine triplet microtubules are embedded in a densely staining cylindrical matrix that is spatially distinct from the pericentriolar material of the centrosome. Structures found in the lumen or core of the centriole include linkers between the triplets, granules, fibers, a cartwheel structure at one end of the centriole, and sometimes a small vesicle. Centrosome An organelle located in the cytoplasm of all animal cells and many plants, fungi, and protozoa that controls the polymerization, position, and polar orientation of many of the cell's microtubules throughout the cell cycle. There is usually one centrosome per cell, located near the cell's center; it doubles during interphase, so there are two when the cell divides. At the onset of mitosis, each centrosome increases the number of microtubules it initiates. These mitotic microtubules are more labile and generally shorter than their interphase counterparts, and as they rapidly grow and shrink they probe the space around the centrosome that initiated them. When the nuclear envelope disperses, the microtubules extend from the centrosome into the former nucleoplasm where the chromosomes have already condensed. Some of these microtubules attach to the chromosomes, while others interact with microtubules produced by the sister centrosome, forming a mitotic spindle that organizes and segregates the chromosomes. During anaphase, sister centrosomes are forced apart as the spindle elongates, allowing each daughter cell to receive one centrosome to organize its microtubules in the next cell generation. The shapes of centrosomes differ widely between organisms. The centrosomes of animal cells usually contain a pair of perpendicular centrioles including a parent centriole formed in an earlier cell generation and a daughter centriole formed during the most recent interphase. Centrioles can serve as basal bodies for the initiation of a cilium or flagellum in the cells that make them. They appear to be essential for the formation of these motile appendages, so centriole inheritance by both daughters at cell division is analogous to the transmission of a gene. |
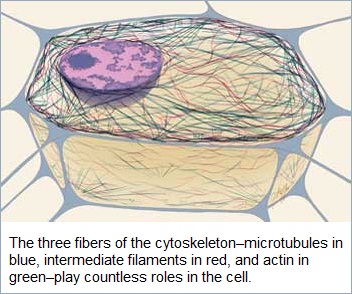
Microtubules can be viewed as part of an overall 3-to-1 arrangement of the Cytoskeleton, though reference to "four" major cytoskeletal filaments appears to be a common expression, though one might argue for another pattern such as 2 X 2 or 2 X 1 X 1, depending on how one cares to characterize structure and or functionality. However, instead of using enumeration, one might opt for words such as singularity, bilarity and trilarity instead of quadrilineal.
Four major types of cytoskeletal filaments are commonly recognized: actin filaments, microtubules, intermediate filaments, and septins. Actin filaments and microtubules are dynamic structures that continuously assemble and disassemble in most cells. Intermediate filaments are stabler and seem to be involved mainly in reinforcing cell structures, especially the position of the nucleus and the junctions that connect cells. Septins are involved in cell division and have been implicated in other cell functions. A wide variety of accessory proteins works in concert with each type of filament, linking filaments to one another and to the cell membrane and helping to form the networks that endow the Cytoskeleton with its unique functions. Many of these accessory proteins have been characterized, revealing a rich diversity in the structure and function of the Cytoskeleton. Source: "Cell." Encyclopædia Britannica Ultimate Reference Suite, 2013. |
Note: by adding "septins", we have created a 3 to 1 ratio.
|
Cytoskeletal elements and Architecture The existence of an organized fibrous array or cytoskeleton in the structure of the protoplasm was postulated in 1928 by Koltzoff. The cytoskeleton can be defined as a cytoplasmic system of fibers which is critical to cell motility. It is dynamic three-dimensional scaffolding contained within a cell's cytoplasm and is made of protein. The ability of eukaryotic cells to adopt a variety of shapes and to carry out coordinated and directed movements depends on the cytoskeleton. The cytoskeleton was known to be unique to eukaryotic cells. Recent research has found cytoskeletal elements in bacteria showing that it has evolved early in evolution. Several proteins that are involved in cell division, cell structure and DNA partitioning have been found to form highly dynamic ring structures or helical ?laments underneath the cell membrane or throughout the length of the bacterial cells. The cytoskeleton can also be referred to as cytomusculature, because, it is directly involved in movements such as crawling of cells on a substratum, muscle contraction and the various changes in the shape of a developing vertebrate embryo; it also provides the machinery for cyclosis in cytoplasm. The main proteins that are present in the cytoskeleton are tubulin (in the microtubules), actin, myosin, tropomyosin and other (in the microfilaments) and keratins, vimenti , desmin, lamin and others (in intermediate filaments). Tubulin and actin are globular proteins, while subunits of intermediate filaments are fibrous proteins. The use of high-voltage electron microscopy on whole cells has helped to demonstrate that there is a highly structured, three-dimensional lattice in the ground cytoplasm. Figure 1 gives an overview of the cytoskeletal system. The primary types of fibers comprising the cytoskeleton are:
They are classified based on their size, function and distribution within the cell. The differences among the three cytoskeletal elements is given in Table 1 and are individually explained in the following subsections.
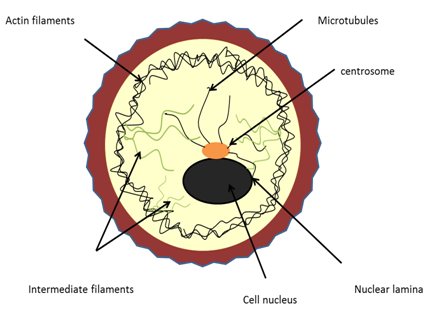 Figure 1: The cytoskeletal system 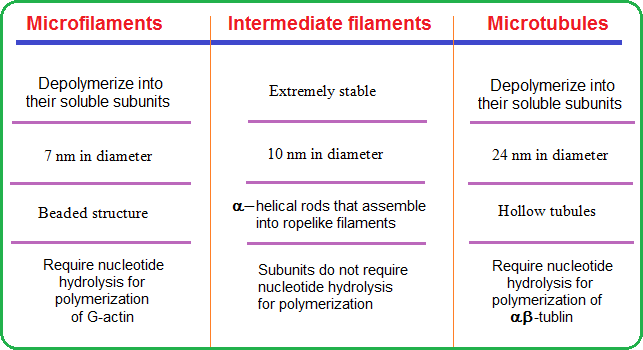 Table 1: Differences among cytoskeletal elements |
Biology 301: Human Physiology, Cell Structure and Function
Page initially created: Friday, 4-August-2017... 4:37 AM
Initial Posting: Thursday, 07-Sept-2017... 4:13 AM
Updated Posting: Sunday, 13th-May-2018... 6:50 AM
Most Recent update: Friday, 17th, March 2023... 10:5 AM
Herb O. Buckland
herbobuckland@hotmail.com